USAGE
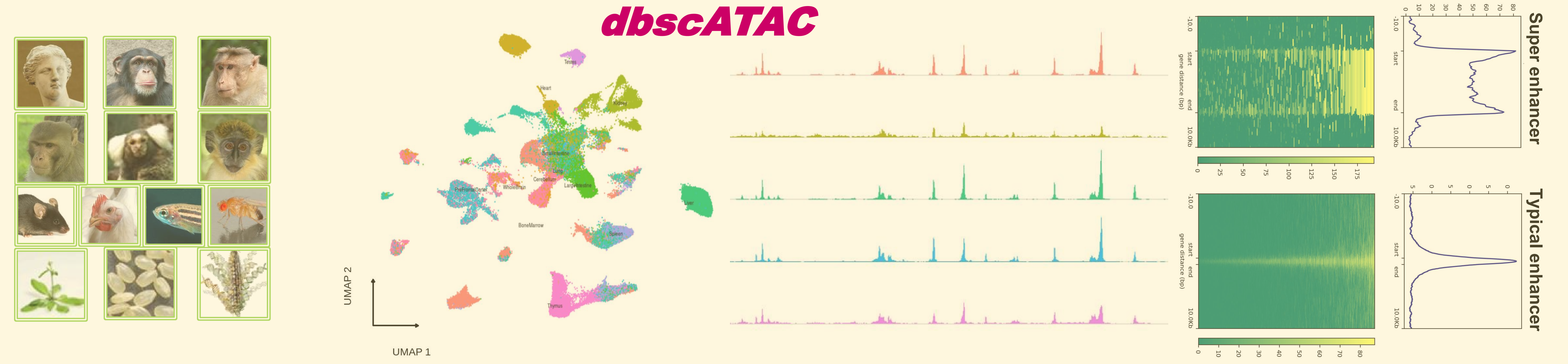
dbscATAC is a single-cell ATAC-seq database with super-enhancer, enhancer, and gene marker annotations from 1028 clustered tissue/cell types in 13 species. Based on the features of single cell super-enhancers, enhancers, or enhancer-gene interactions, a user-friendly platform with eight useful modules for searching, visualizing, and browsing super-enhancers, enhancers, and gene markers are designed.
1. Search single-cell superenhancers:: By selecting a species and providing a cell/tissue type, users are able to identify the single-cell superenhancers in this cell type of selected species.
2. Search single-cell gene markers:: By selecting a species and providing a cell/tissue type, users can search for the single-cell gene markers in the selected cell type of species.
3. Search single-cell enhancers:: By selecting a species and tissue/cell type, users can search the single-cell enhancers for this tissue/cell type.
4. Display the differences among tissue/cell types by single-cell enhancer clustering:: Users can analyze the differences of scATAC-seq data among different tissue/cell types by clustering with selected cell types.
5. Identification of tissue/cell type-specific superenhancer/enhancers at the single-cell level:: Users can identify the tissue/cell type-specific superenhancer/enhancers at the single-cell level with selected cell types.
6. Compare single-cell enhancers across different tissue/cell types with a given region:: Users Search and compare the single-cell enhancer distributions across selected tissue/cell types with a given region.
7. Compare single-cell enhancers of a given gene in different tissue/cell types: Given a gene, users can find and compare its enhancers across selected tissue/cell types.
8. Predict the single-cell cis-regulatory elements and relative target genes for a list of given genomic regions: Users can predict the enhancer or promoter properties and even their target genes for the input genomic regions.