Download
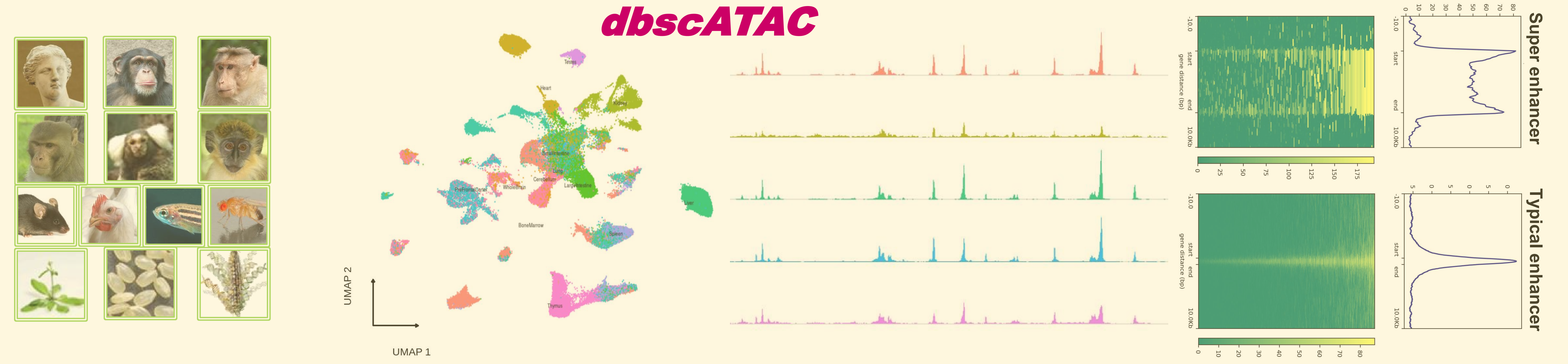
dbscATAC utilized a set of improved machine learning algorithms to identify super-enhancers and gene markers in hundreds of thousands of single cells. 155,942 super-enhancers, 288,480 marker genes, 11,900,573 enhancers and 8,986,510 enhancer-gene interactions from 1,381,777 single cells across 881 tissue/cell types in 13 species were identified. The information of all individual datasets in each tissue/cell type was also attached.
※Single-cell super-enhancers:
※Single-cell marker genes:
| Axillary_bud_II | Leaf | Seedling | Root_II | Root_I |
| .. | Root_Crown_I | Ear | . | Root_Crown_II |
| Tassel | Axillary_bud_I |
※Single-cell enhancers:
| Tassel | Axillary_bud_II | Root_Crown_II | Root_Crown_I | Ear |
| .. | . | Seedling | Leaf | Root_I |
| Axillary_bud_I | Root_II |
| .. | . | Neur_crest |
※Enhancer-gene interactions at the single-cell level:
| Seedling | Axillary_bud_I | Root_I | Axillary_bud_II | Root_Crown_I |
| .. | . | Root_Crown_II | Leaf | Ear |
| Tassel | Root_II |
| Neur_crest | .. | . |
※SNPs within single-cell enhancers:
※TF binding motifs within single-cell enhancers:
※Disease enhancers annotated by HEDD: