dbscATAC
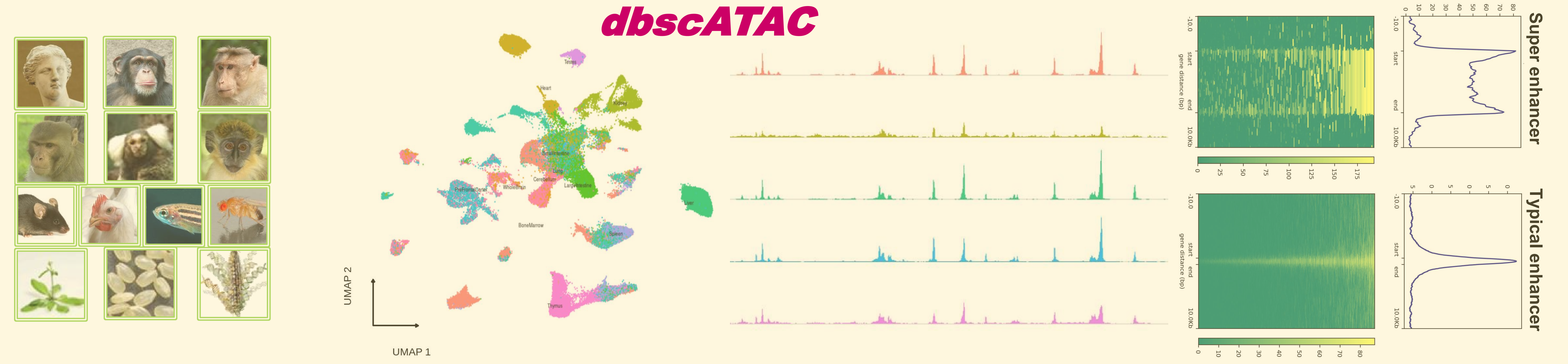
The database provides single-cell ATAC-seq annotations in thriteen species, including human (hg38), chimp (panTro5), rhesus (rheMac10), cynomolgus (macFas6), vervet (ChlSab1.1), marmoset (calJac3), mouse (mm10), chicken (galGal6), zebrafish (danRer10), fly (dm6), rice (IRGSP1), arabidopsis (TAIR10), and maize (B73v4). The consensus enhancers were identified based on single-cell datasets of scATAC-seq. Currently, dbscATAC contains 213,835 super-enhancers, 347,484 marker genes, 13,470,526 enhancers and 10,402,346 enhancer-gene interactions from 1,668,076 single cells across 1,028 cluster/cell types in thirteen species. The database allows users to search the super-enhancers, marker genes, and enhancers in simple and advanced ways. Advanced Search: (1) Display the differences among tissue/cell types by single-cell enhancer clustering; (2) Identification of tissue/cell type-specific superenhancer/enhancers at the single-cell level; (3) Compare single-cell enhancers across different tissue/cell types with a given region; (4) Compare single-cell enhancers of a given gene in different tissue/cell types; (5) Predict the cis-regulatory elements and relative target genes for a list of given genomic regions.
Advanced Search
※ Display the differences among tissue/cell types by single-cell enhancer clustering (batch information):
※ Identification of tissue/cell type-specific superenhancer/enhancers at the single-cell level (batch information):
※ Compare single-cell enhancers across different tissue/cell types with a given region:
※ Compare single-cell enhancers of a given gene in different tissue/cell types:
※ Predict the single-cell cis-regulatory elements and relative target genes for a list of given genomic regions: